| biplot.psych {psych} | R Documentation |
Draw biplots of factor or component scores by factor or component loadings
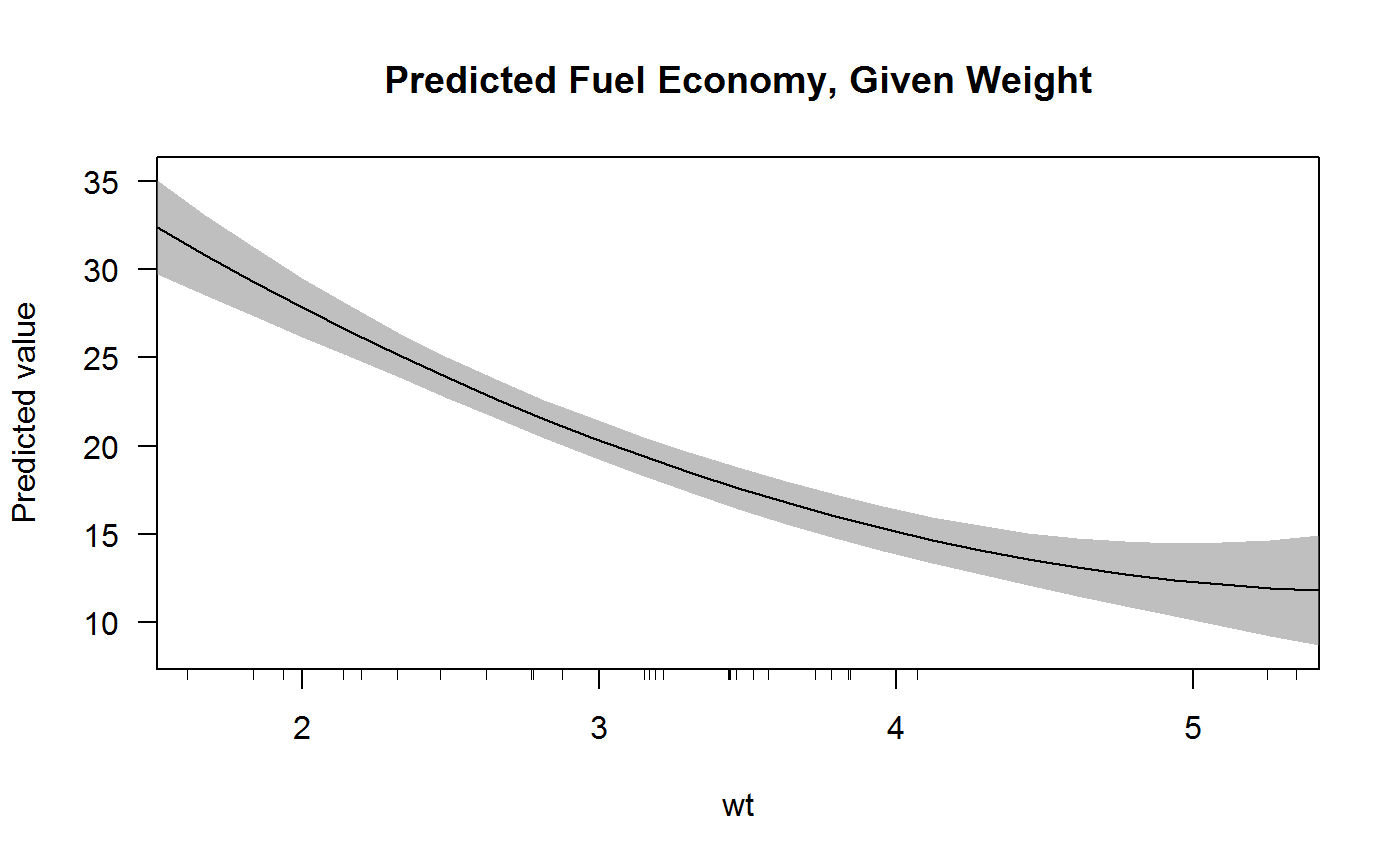
Creating a Factor. The command used to create or modify a factor in R language is – factor with a vector as input. The two steps to creating a factor are: Creating a vector; Converting the vector created into a factor using function factor Examples: Let us create a factor gender with levels female, male and transgender. How to make interactive 3D scatter plots in R. Building AI apps or dashboards in R? Deploy them to Dash Enterprise for hyper-scalability and pixel-perfect aesthetic. 10% of the Fortune 500 uses Dash Enterprise to productionize AI & data science apps. Use the default method - plot.default while turning the axes off. Then use the axis function to create custom axes. I used a 1/20 scale for my means before using them to set cex. R - Factors - Factors are the data objects which are used to categorize the data and store it as levels. They can store both strings and integers. They are useful in the colu. Because R does not impose this restriction and further makes subsetting expressions very simple, that feature is not really useful and can be achieved using standard subsetting notation in R: x factor (cyl). am + hp + wt, data = mtcars) # automatic vehicles margins (x, data = mtcarsmtcars $ am 0, ).
Description
Extends the biplot function to the output of fa, fa.poly or principal. Will plot factor scores and factor loadings in the same graph. If the number of factors > 2, then all pairs of factors are plotted. Factor score histograms are plotted on the diagonal. The input is the resulting object from fa, principal, or }code{linkfa.poly with the scores=TRUE option. Points may be colored according to other criteria.
Usage
Arguments
x | The output from |
labels | if NULL, draw the points with the plot character (pch) specified. To identify the data points, specify labels= 1:n where n is the number of observations, or labels =rownames(data) where data was the data set analyzedby the factor analysis. |
cex | A vector of plot sizes of the data labels and of the factor labels |
main | A main title for a two factor biplot |
hist.col | If plotting more than two factors, the color of the histogram of the factor scores |
xlim.s | x limits of the scores. Defaults to plus/minus three sigma |
ylim.s | y limits of the scores.Defaults to plus/minus three sigma |
xlim.f | x limits of the factor loadings.Defaults to plus/minus 1.0 |
ylim.f | y limits of the factor loadings.Defaults to plus/minus 1.0 |
maxpoints | When plotting 3 (or more) dimensions, at what size should we switch from plotting 'o' to plotting '.' |
adjust | an adjustment factor in the histogram |
col | a vector of colors for the data points and for the factor loading labels |
pos | If plotting labels, what position should they be in? 1=below, 2=left,3 top, 4 right.If missing, then the assumption is that labels should be printed instead of data points. |
arrow.len | the length of the arrow head |
pch | The plotting character to use. pch=16 gives reasonable size dots.pch='.' gives tiny points. If adding colors, use pch between 21 and 25. (see examples). |
choose | Plot just the specified factors |
cuts | Do not label cases with abs(factor scores) < cuts) (Actually,the distance of the x and y scores from 0) |
cutl | Do not label variables with communalities in the two space < cutl |
group | A vector of a grouping variable for the scores. Show a different color and symbol for each group. |
... | more options for graphics |
Details
Uses the generic biplot function to take the output of a factor analysis fa, fa.poly or principal components analysis principal and plot the factor/component scores along with the factor/component loadings.
This is an extension of the generic biplot function to allow more control over plotting points in a two space and also to plot three or more factors (two at time).
This will work for objects produced by fa, fa.poly or principal if they applied to the original data matrix. If however, one has a correlation matrix based upon the output from tetrachoric or polychoric, and has done either fa or principal on the correlations, then obviously, we can not do a biplot. However, both of those functions produce a weights matrix, which, in combination with the original data can be used to find the scores by using factor.scores. Since biplot.psych is looking for two elements of the x object: x$loadings and x$scores, you can create the appropriate object to plot. See the third example.
Author(s)
William Revelle
See Also
fa, fa.poly, principal, fa.plot, pairs.panels
Examples
Hi
I have a large data frame of gene sets whose components are in the form of gene symbols. I'm trying to use biomaRt to convert these symbols into entrez IDs so that I can run the sets through MAGMA. However, I can't work out how to convert the data frame without destroying the structure of the gene sets. I always get a data frame just listing the the IDs of genes in my sets, which obviously isn't very useful for me. Can anyone help?
I thought this would work to preserve the headers of the original data frame but I still just get a list:
Thanks
Merge by gene symbols with your data frame. getBM function ouput is data frame with gene symbol and NCBI gene ID. Now merge both the data frames by gene symbol. If you do not know how to merge data frame, post example data here from big data frame.
Thanks! But yeah I don't know how to merge them by gene symbol. Can you direct me to any useful webpages or packages that teach this?
Mt data set looks like this:
But formatted by rows, the sets are a lot longer and there's about 1600 of them.
Thanks
could you please reformat your post, so that data frame can be clear? You can look at the merge examples here: https://www.statmethods.net/management/merging.html
Plot With Factors Review
Looks like genomax reformatted it for me. Thanks genomax
For future posts: you can do this formatting by selecting the text and clicking the 101010 button. When you compose or edit a post that button is in your toolbar, see image below:
I am assuming every row has different number of genes?
Hi
Cplot With Factors R
I am new to this R and Bioinformatics game. I tried using the code posted by cpad0112 with my dataset and got the following error.
Plot With Factors In R
I think I have found a workaround using 'list attributes' to find alternative attributes but would appreciate it somebody could confirm what attribute people use in place of hngc_symbol or correct any errors in my code.
Plot With Factors Ratio
thanks
Peter
Plot With Factors Reading
Plot With Factors Ratios
Please use ADD COMMENT/ADD REPLY when responding to existing posts to keep threads logically organized.SUBMIT ANSWER is for new answers to original question.